Compound identification in metabolomics
Jessica Cooperstone
How do we go from features to metabolites?
- Individually (one feature at a time)
- Manual, very high confidence in ID
- In bulk (many features at a time)
- Computational, generally lower confidence in ID
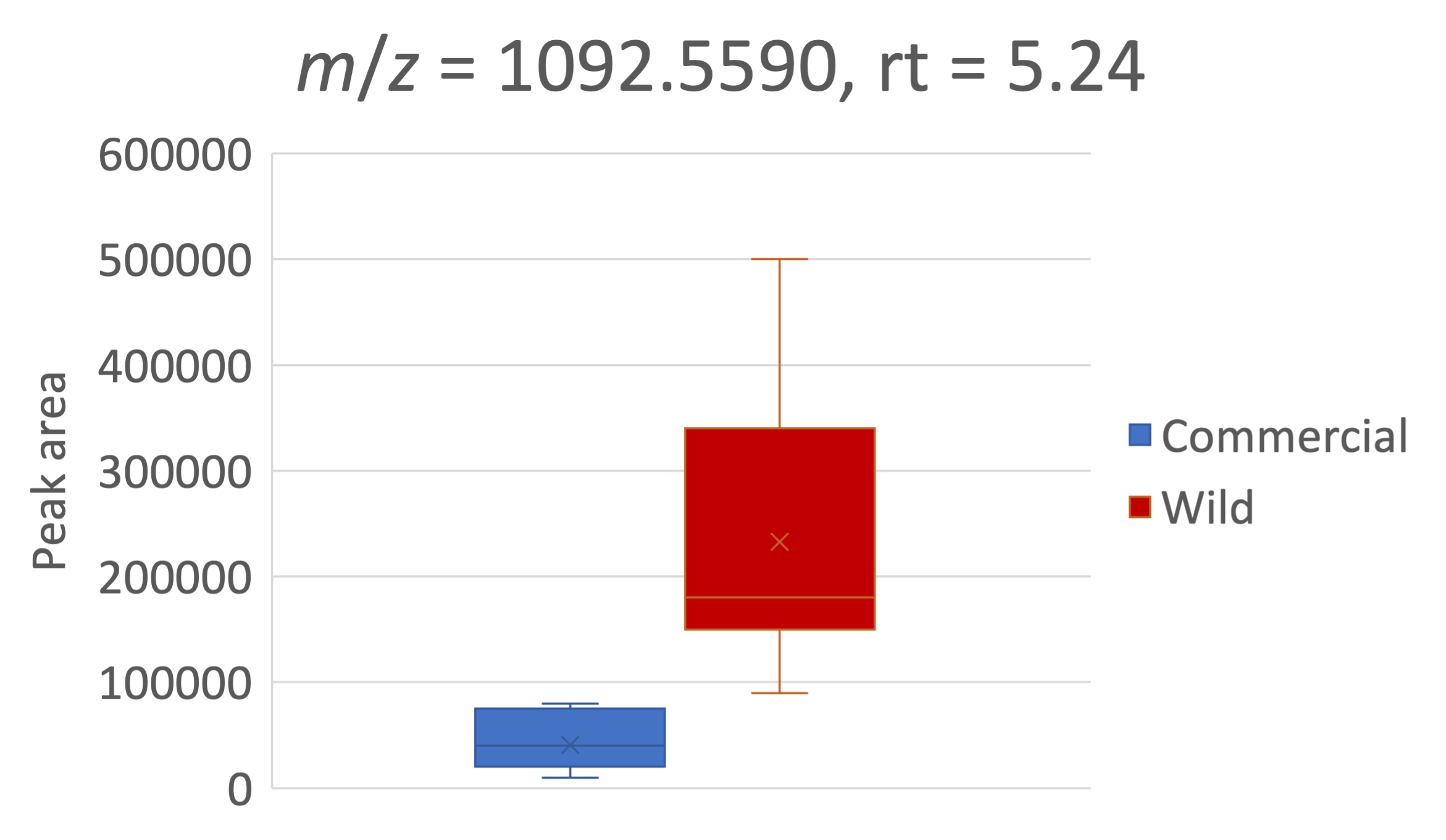
Starting with a m/z
- A feature of interest differentially present in commercial vs. wild tomatoes. We want to know what this is.
- UHPLC-QTOF-MS, ESI+, reversed phase C18, Methanol extract
- A useful adduct calculator
Search in MS databases - HMDB
Search > LC-MS Search
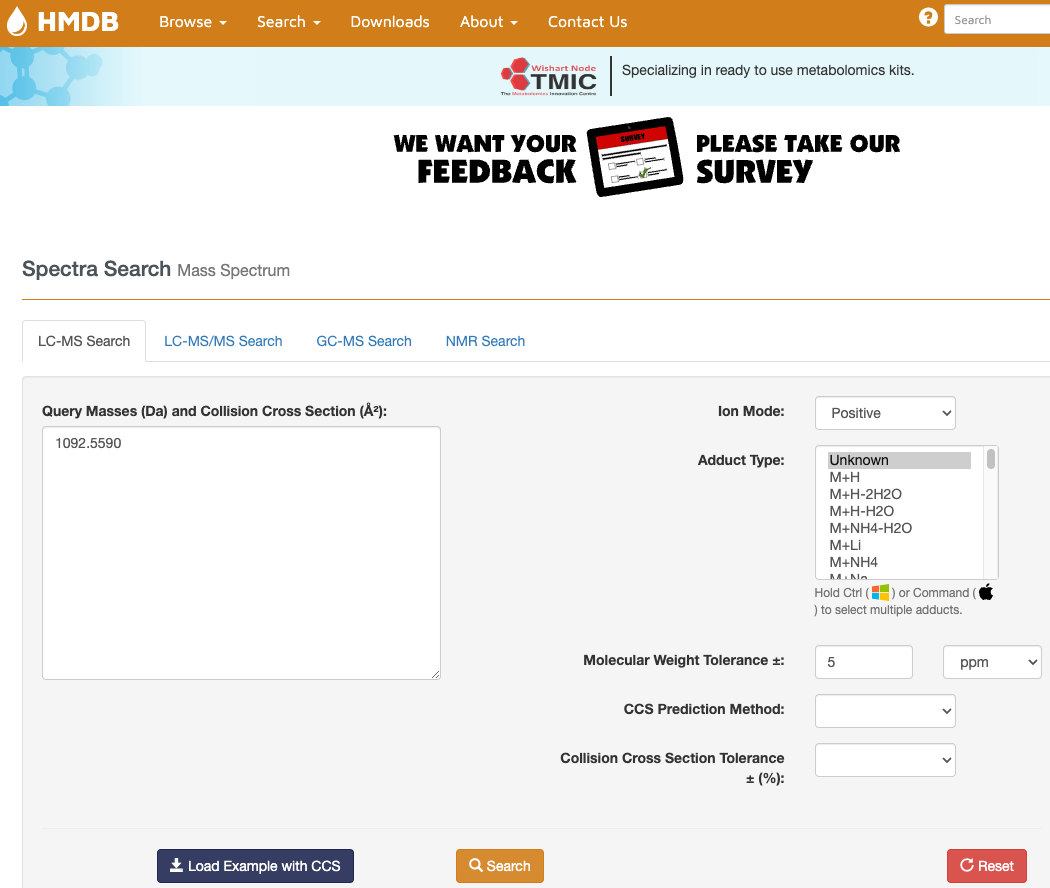
- Enter your mass and mode
- Indicate adduct type (can put unknown if you don’t know)
- Select a mass error (5 ppm is good for a QTOF)
- Search
Evaluate search results
Think about:
- Which structures are plausible?
- Make sense in your biological system
- Make sense in your extraction
- Make sense based on retention time
- Which adducts are more likely?
- The top result is not necessarily your ID!
Access or collection MS/MS fragmentation data

MS/MS spectra of 1092.5590 at 65eV in +ESI
Download structure
http://www.hmdb.ca/spectra/ms_ms/59530
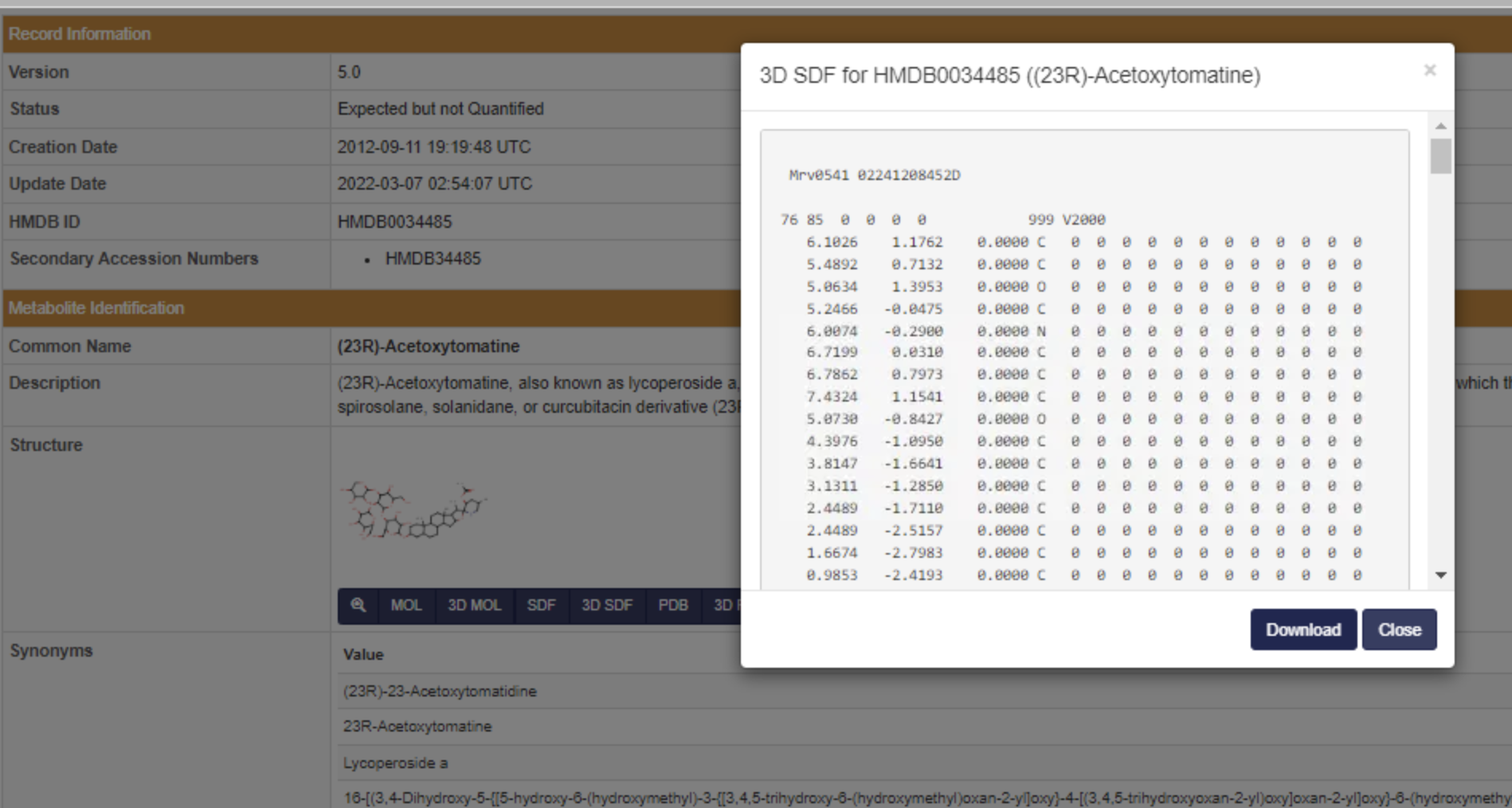
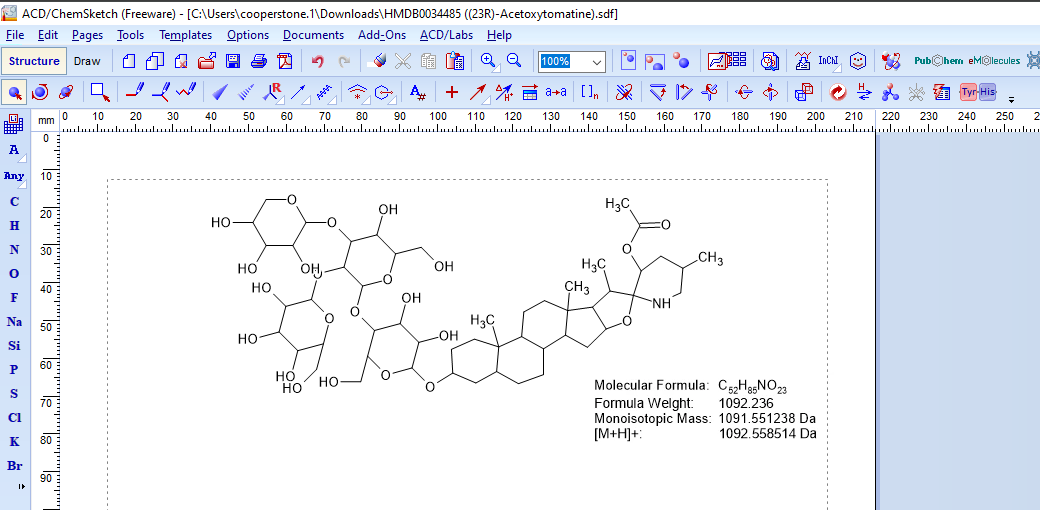
Download an .sdf file of your structure
Import into ChemSketch
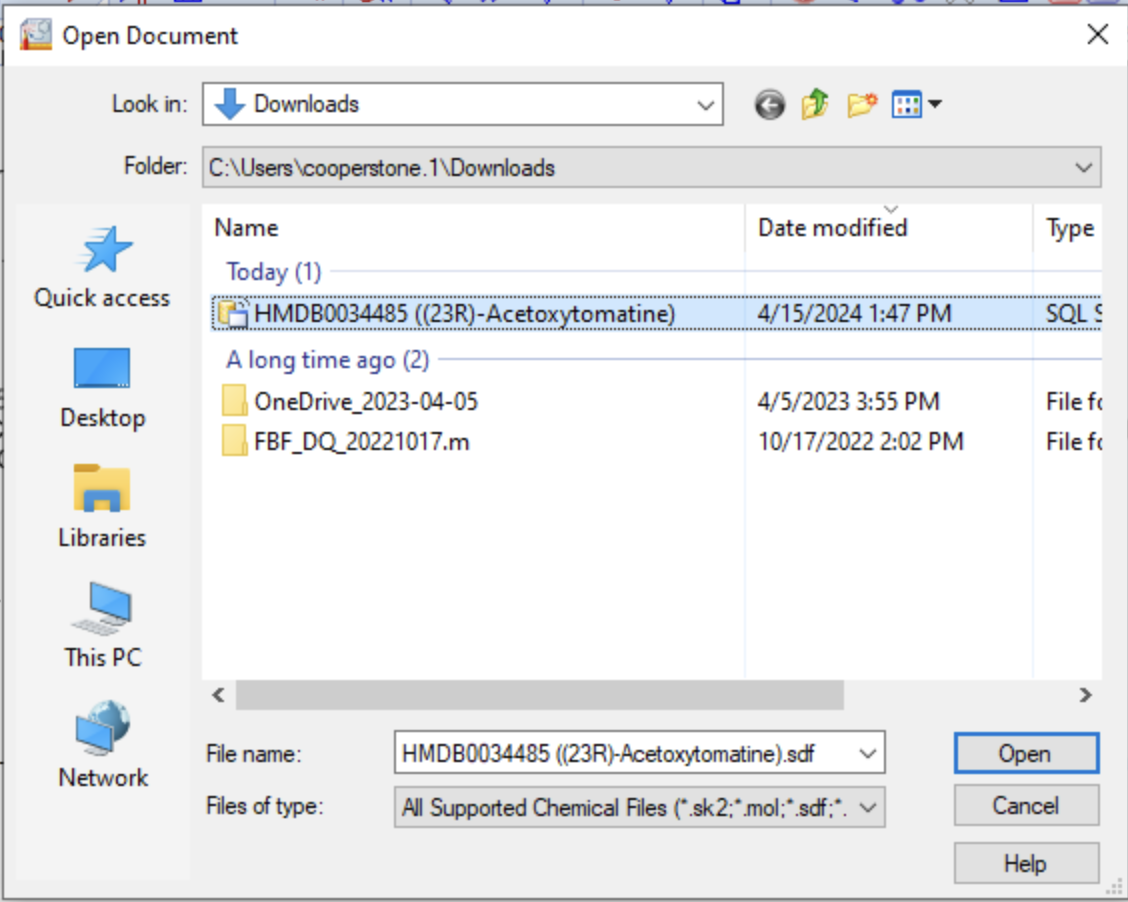
Import your .sdf file into ChemSketch
Set ChemSketch to provide you structural information
- Select which parameters you want printed:
Tools>Calculate>Select properties to calculate> Select “Molecular Formula”, “Monoisotopic Mass”, “[M+H]+” - Have ChemSketch calculate those parameters:
Tools>Calculate>Selected properties>Copy to editor.
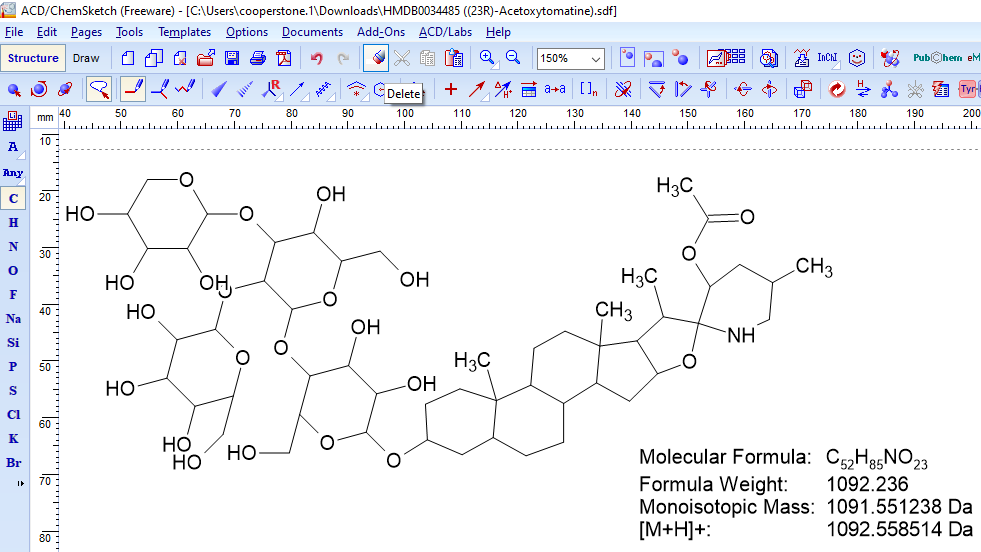
Breaking bonds to rationalize fragments
- Use the eraser (delete) to break bonds (learn more here re: using ChemSketch)
- Select a bond and it will be deleted
- The details will stay with the larger fragment. Highlight the smaller piece to recalculate parameters.
- Account for any rearrangement or additions/subtractions
Breaking bonds to rationalize fragments
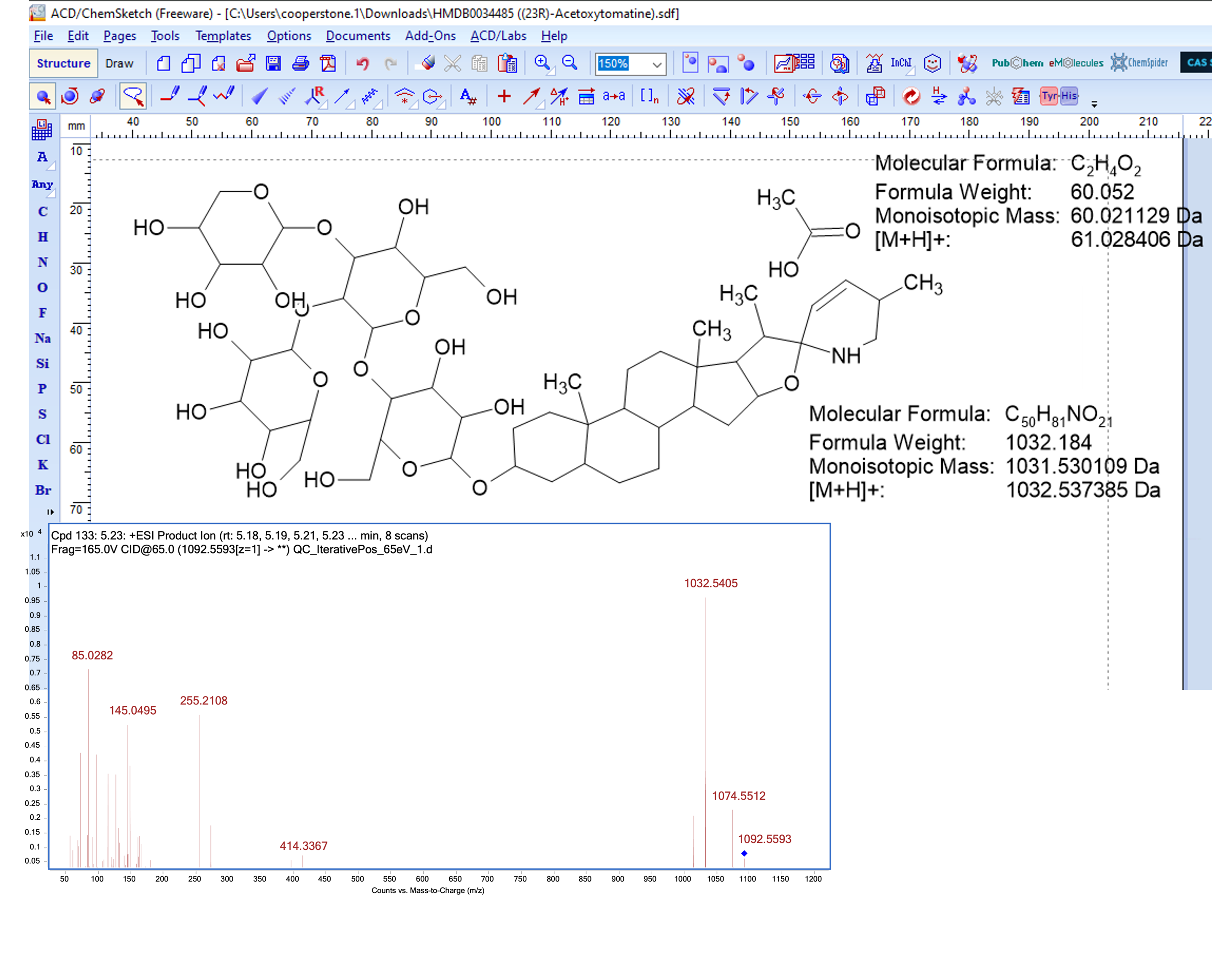
Cleaving at the acetoxy group
Breaking bonds to rationalize fragments
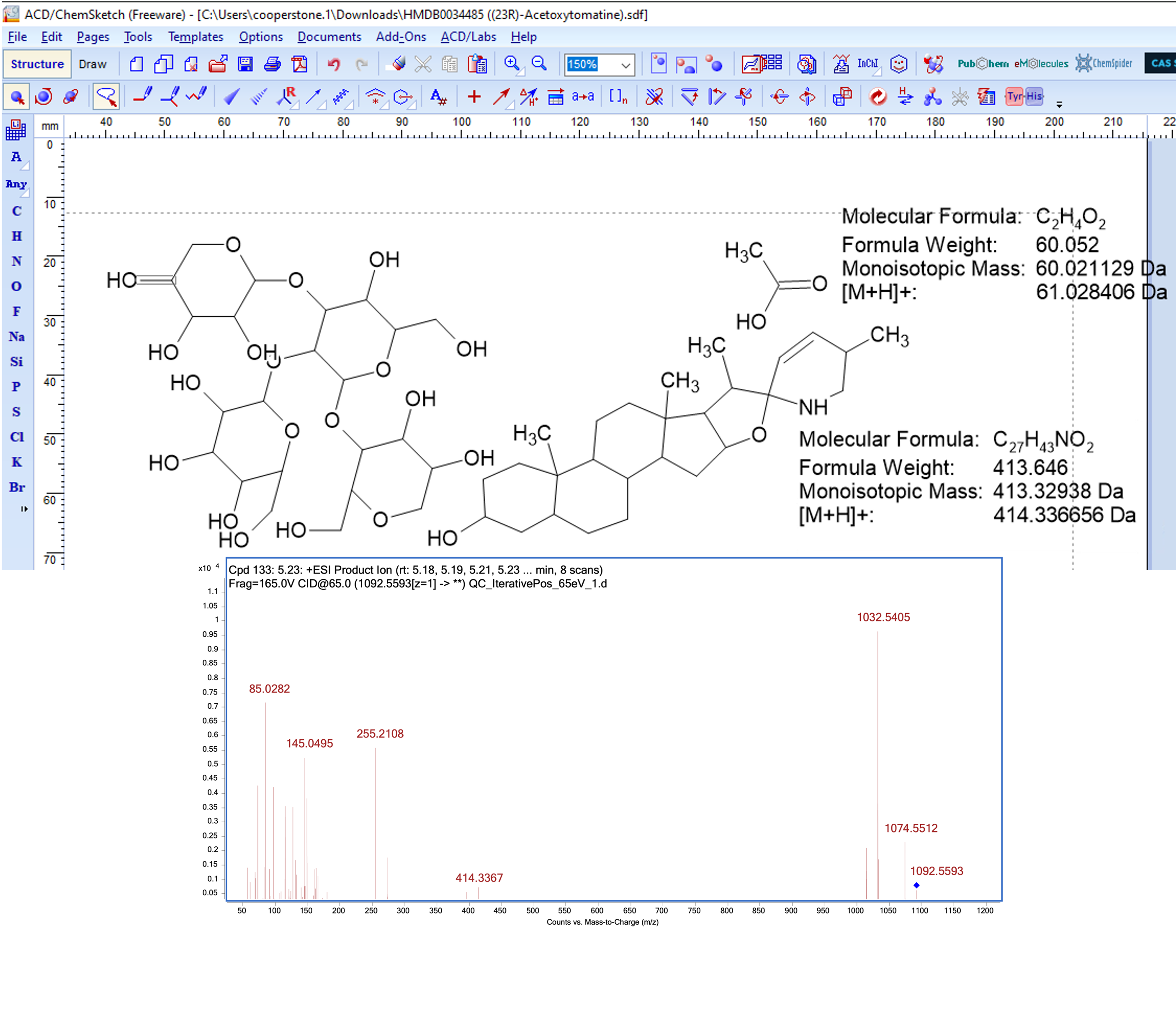
Cleaving at the acetoxy group
How can we improve the confidence of our ID?
- Reference against publicly available MS/MS spectra
- Purchase authentic standard and compare
- Synthesize standard, confirm by NMR, and hope you’re right 🥹
- Compare to MS/MS spectra of similar compounds
- Compare to a sample that you know has your compound of interest
MS/MS online databases
- Experimental spectra
- Predicted spectra
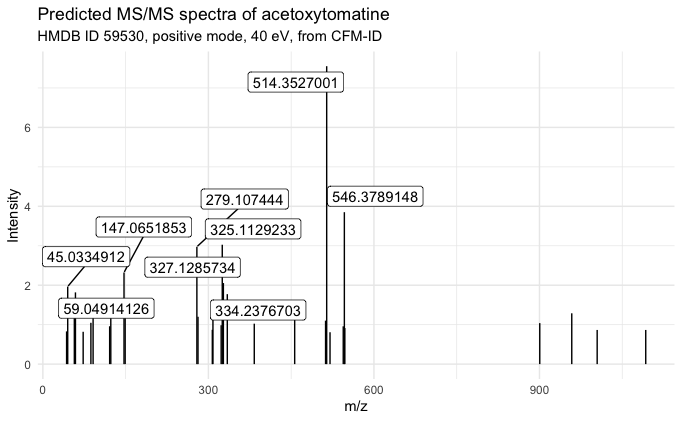
MS/MS predicted spectra of acetoxytomatine
http://www.hmdb.ca/spectra/ms_ms/59530
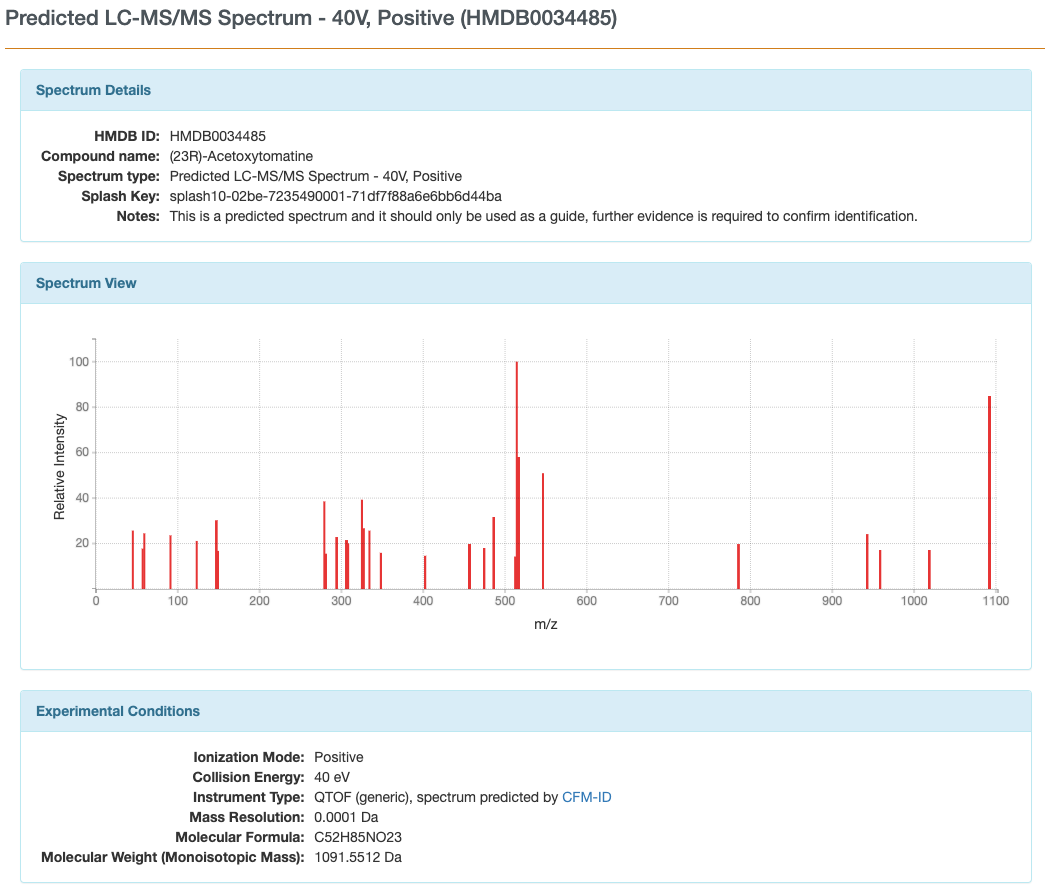
Predicted spectra for (23R)-acetoxytomatine
Predicted vs. actual

MS/MS spectral databases
- GNPS
- MassBank of North America (MONA)
- HMDB
- Others that you have to pay for (METLIN, NIST, mzCloud)
- A review on spectral libraries by Bittremieux et al., Metabolomics 2022.
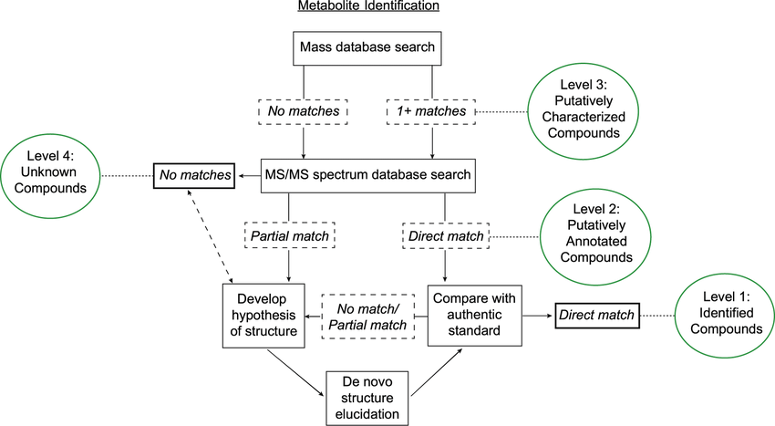
LC-MS metabolite ID workflow
Do you need to have an ID to quantify?
© Jessica Cooperstone, 2024